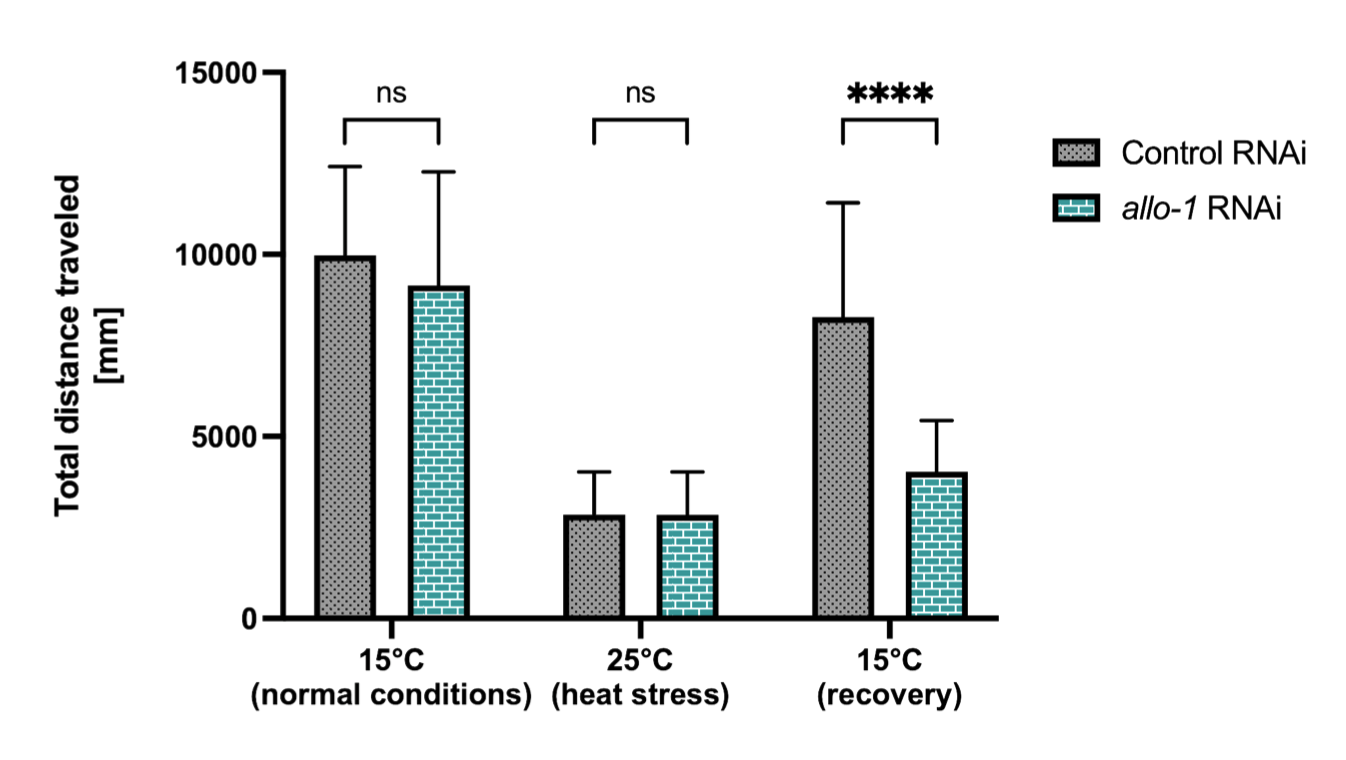
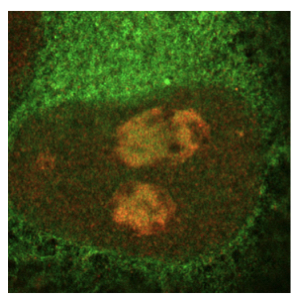
The ubiquitin-conjugating enzyme UBE2K determines neurogenic potential through histone H3 in human embryonic stem cells
Fatima, A., Irmak, D., Noormohammadi, A., Rinschen, M.M., Das, A., Leidecker, O., Schindler, C., Sánchez-Gaya, V., Wagle, P., Pokrzywa, W., Hoppe, T., Rada-Iglesias, A., & Vilchez, D.
Communications Biology
doi: 10.1038/s42003-020-0984-3
Ubiquitin signaling regulates RNA biogenesis, processing, and metabolism
Thapa, P., Shanmugam, N., & Pokrzywa, W.*
BioEssays
doi: 10.1002/bies.201900171
CHIP ubiquitylates NOXA and induces its lysosomal degradation in response to DNA damage
Albert, M.-C., Brinkmann, K., Pokrzywa, W., Günther, S. D., Krönke, M., Hoppe, T., & Kashkar, H.
Cell Death & Disease
doi: 10.1038/s41419-020-02923-x
Bioshell 3.0: Library for processing structural biology data
Macnar, J.M., Szulc, N.A., Kryś, J.D., Badaczewska-Dawid, A.E., & Gront, D.
Biomolecules
doi: 10.3390/biom10030461
Pathogenic variants in the myosin chaperone UNC-45B cause progressive myopathy with eccentric cores
Donkervoort, S., Kutzner, C. E., Hu, Y., Lornage, X., Rendu, J., Stojkovic, T., Baets, J., Neuhaus, S.B., Tanboon, J., Maroofian, R., Bolduc, V., Mroczek, M., Conijn, S., Kuntz, N. L., Töpf, A., Monges, S., Lubieniecki, F., McCarty, R. M., Chao, K. R., Governali, S., Böhm, J., Boonyapisit, K., Malfatti, E., Sangruchi, T., Horkayne-Szakaly, I., Hedberg-Oldfors, C., Efthymiou, S., Noguchi, S., Djeddi, S., Iida, A., di Rosa, G., Fiorillo, C., Salpietro, V., Darin, N., Faure, J., Houlden, H., Oldfors, A., Nishino, I., de Ridder, W., Straub, V., Pokrzywa, W., Laporte, J., Foley, R., Romero, N.B., Ottenheijm, C., Hoppe, T., & Bönnemann, C.G.
The American Journal of Human Genetics
doi: 10.1016/j.ajhg.2020.11.002
---
bolded - member of the Pokrzywa lab
* corresponding author